RESEARCH
The research programme can be divided into three different yet interconnected areas:
Chemical biology
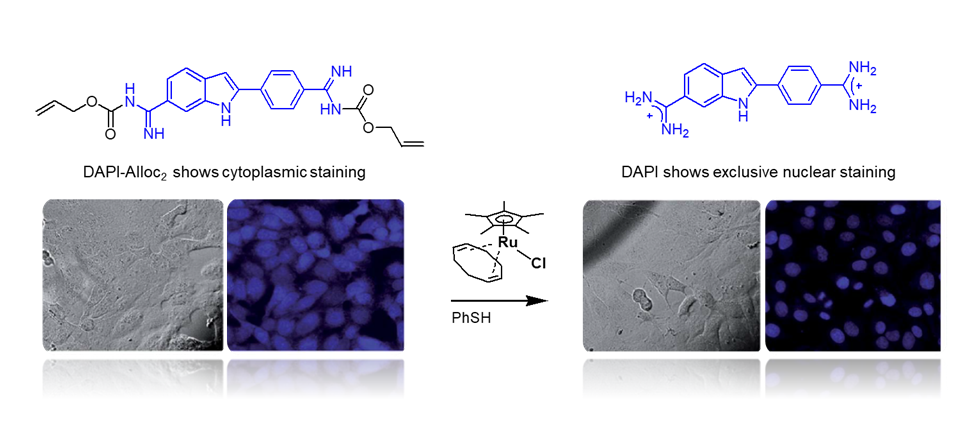
Sanchez MI et al. Chem. Sci. 2014, 10, 8668.
Protein engineering
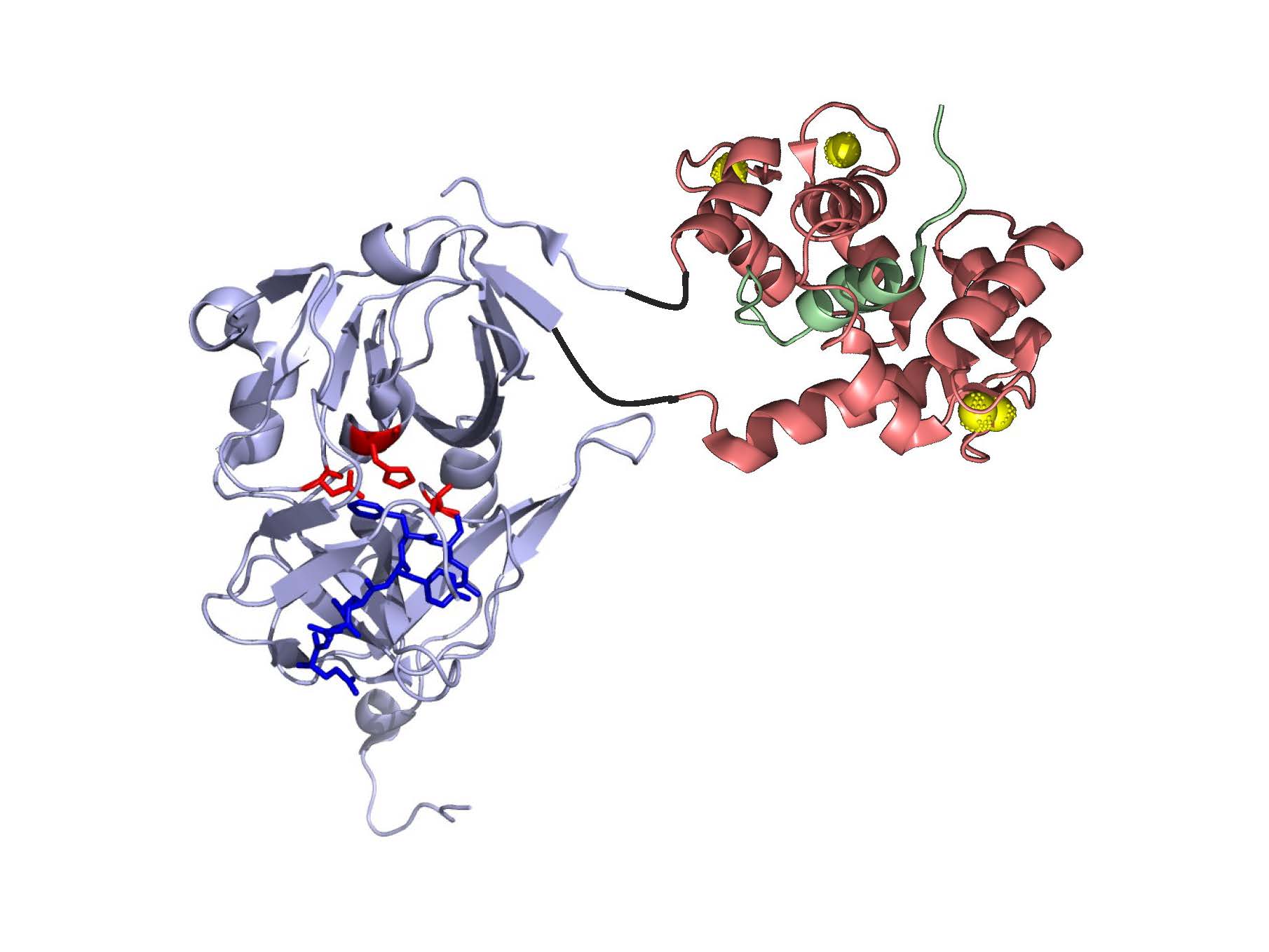
Model of the best CaTEV design based on crystal structure of wild-type TEV protease and a calmodulin complex. (PNAS 2020, 117, 33186)
Protein engineering for cell biology and neuroscience: The ability to create and modify the functional properties of a specific protein is key for the development of molecular biotechnology and has significant potential for application in various fields. These protein hybrids have been the basis of the development of new molecular tools such as scFLARE (for single-chain Fast Light- and Activity-Regulated Expression) or FLiCRE (for Fast Light and Calcium-Regulated Expression). For example, scFLARE relies on an engineered LOV domain–caged protease cleavage site and a calcium-activated TEV protease. (Sanchez MI et al. PNAS 2020, 117, 33186) scFLARE has been used in neuroscience to record patterns of neuronal activation with a temporal resolution of minutes. The development of protein chimeras normally conveys a decrease in the natural activity of both parts because the incorporation of the different modules affects the geometry and the overall structure. Directed evolution is a very powerful method in protein engineering as it mimics the natural process of Darwinian evolution and is one of the core capabilities of the lab.
For instance, TEV protease (Tobacco Etch Virus protease) is one of the most widely used proteases in biotechnology, proteomics, and protein purification to remove affinity tags due to its exquisite sequence specificity. A limitation, however, is its slow catalytic rate. To overcome this limitation, I created a new platform in yeast for the directed evolution of a faster TEV. (Sanchez MI et al. Nat. Methods 2020,17, 164) These new mutants were incorporated into FLiCRE, endowing it with an unprecedented temporal resolution to label and manipulate active neurons underlying specific behaviours. (Kim CK* & Sanchez MI* et al. Cell 2020, 183, 2003). Another advantage of TEV protease is its orthogonality against the mammalian proteome, which makes it the protease of choice for other applications. In collaboration with other scientists, we are using an evolved version of TEV to study the mechanics of the cardiovascular system.
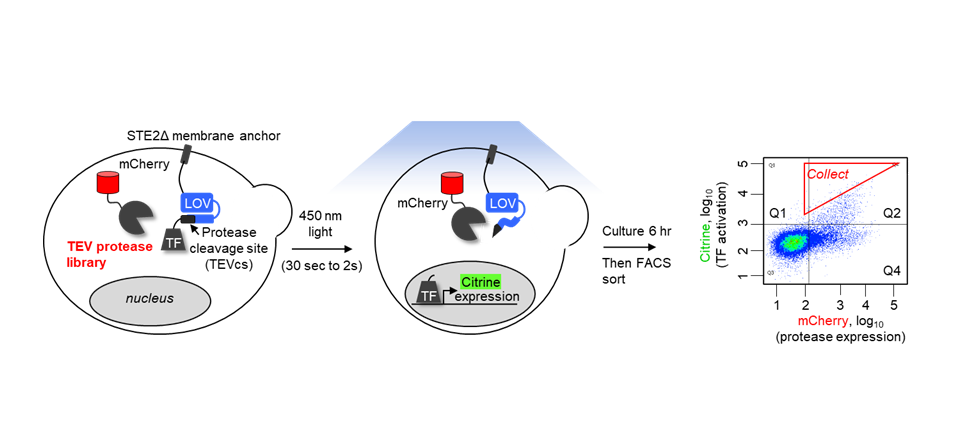
Schematic of the evolution platform to generate TEV protease variants. Catalysis by TEV protease allows a transcription factor to enter the nucleus and modulate transcription of the mCitrine gene. mCherry is constitutively expressed as protease expression marker (Nat. Methods 2020,17, 164).
In ongoing work, we are creating programmable proteases, recombinases, and other enzymes for different purposes while we develop new platforms for directed evolution.
Synthetic biology
Synthetic biology and artificial genomes: Synthetic biology is a new discipline that aims to understand, build, and manipulate the mechanism of life and cellular functions by applying engineering principles. Directed evolution has proven to be key for the improvement of different enzymatic activities which have revolutionised many fields from neuroscience to genome editing. However, more than 90% of published papers working on directed evolution take place in Escherichia coli and Saccharomyces cerevisiae.
Unfortunately, many proteins such as Cytochromes or GPCRs require specific environments, post-transcriptional modifications, and so on, to be active. To address this limitation, we aim to improve the features of different microrganisms and cellular lines such as Chlamydomonas reinhardtii (green algae) or Spodoptera frugiperda Sf9 (insect cell line) to make them suitable host organisms to carry out different evolutionary campaings. We are also starting a new project where we aim to create a synthetic cell bearing a minimun genome to better understand the basic principles and requirements of life.

